
The Fowler Lab

Our Research

Overview
Our research is focused on understanding the biology of bacterial pathogens, how they interact with their hosts and how they acquire new virulence, and disease properties. One major goal of the lab is to identify and characterize genetic factors that enable closely related bacteria (such as members of the same species or subspecies) to exhibit very different traits.
Much of our research is focused on the bacterial species Salmonella enterica. Salmonella is an ideal subject for our research due to its importance to human health and agriculture as well as the fact that it is a very diverse species comprised of many lineages that exhibit very different behaviours and that occupy different ecological niches.
Each area of research in the lab aims to shed light on an interesting and unexplored aspect of biology as well as to build knowledge and tools with real world applications. We are interested in devising new approaches to combat bacterial infections, re-engineering bacterial products into therapeutics, and developing new synthetic biology tools.
Some Examples
Our Publications
Current Featured Publication:
Chemello AJ, Fowler CC. Alternate typhoid toxin assembly evolved independently in the two Salmonella species. Miller SI, ed. mBio. 2024;15(4):e03403-23. doi:10.1128/mbio.03403-23
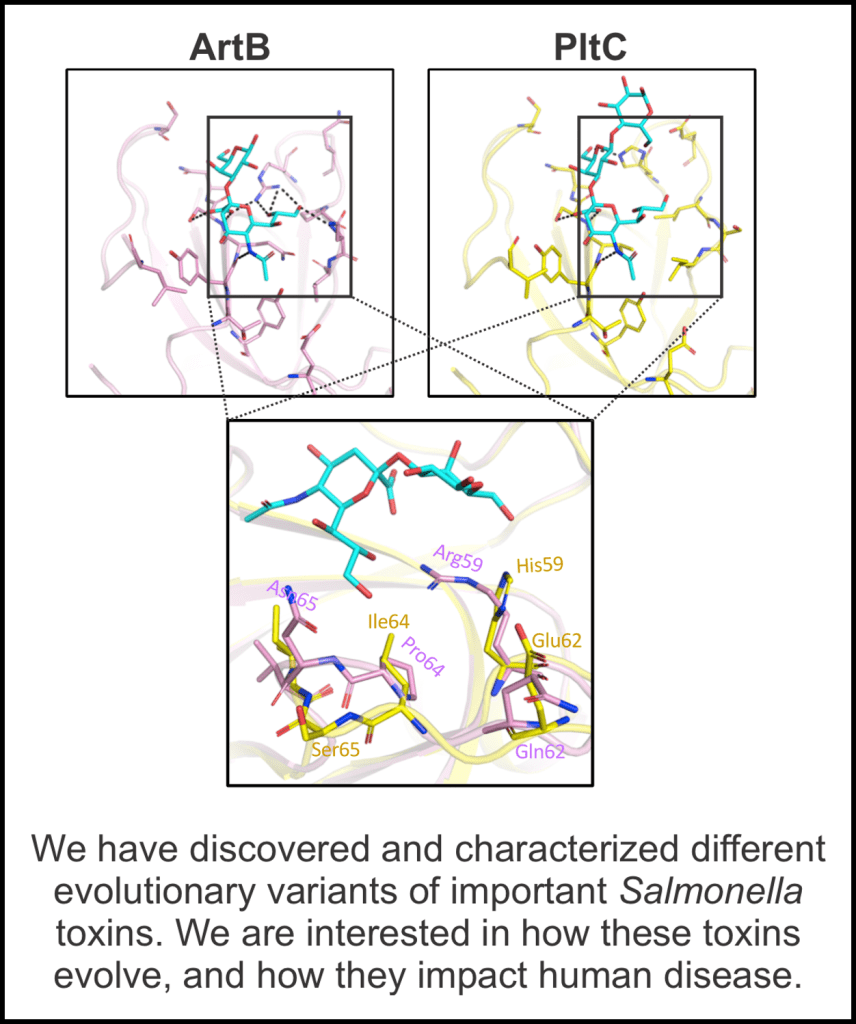
Typhoid toxin is an AB5 toxin produced by many Salmonella sp. bacteria, including S. Typhi, the causative agent of typhoid fever. The toxin is composed of three subunits, two active subunits (CdtB and PltA) and one binding subunit (PltB). In this paper, we show that two distinct typhoid toxins are produced in Salmonella bongori. S. bongori incorporates not only PltB into typhoid toxin, but also an alternative binding subunit, PltD. We found that PltD typhoid toxin is less toxic to human epithelial cells than PltB typhoid toxin. This exemplifies how bacterial toxins can recruit alternative binding subunits to expand/modify their function.
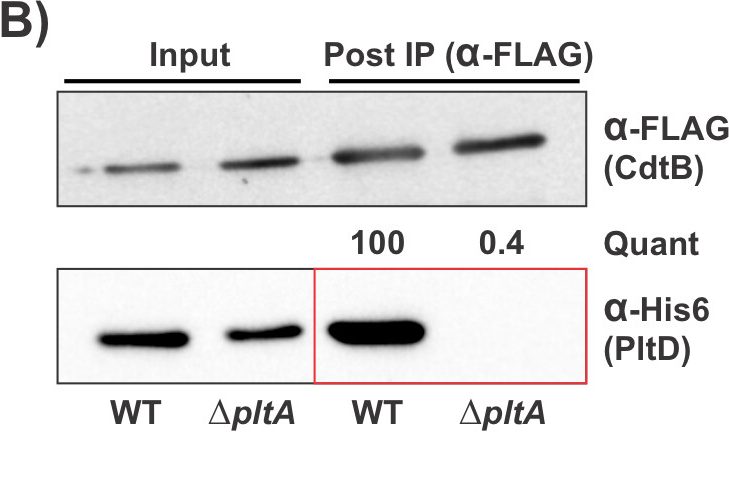
This figure demonstrates that PltD is associated with CdtB in a PltA dependent manner in an immunoprecipitation experimentation with S. bongori.

This figure demonstrates that PltD typhoid toxin is less toxic to human cells than PltB typhoid toxin in toxicity assays using S. bongori deletion strains.
Representative Recent Publications
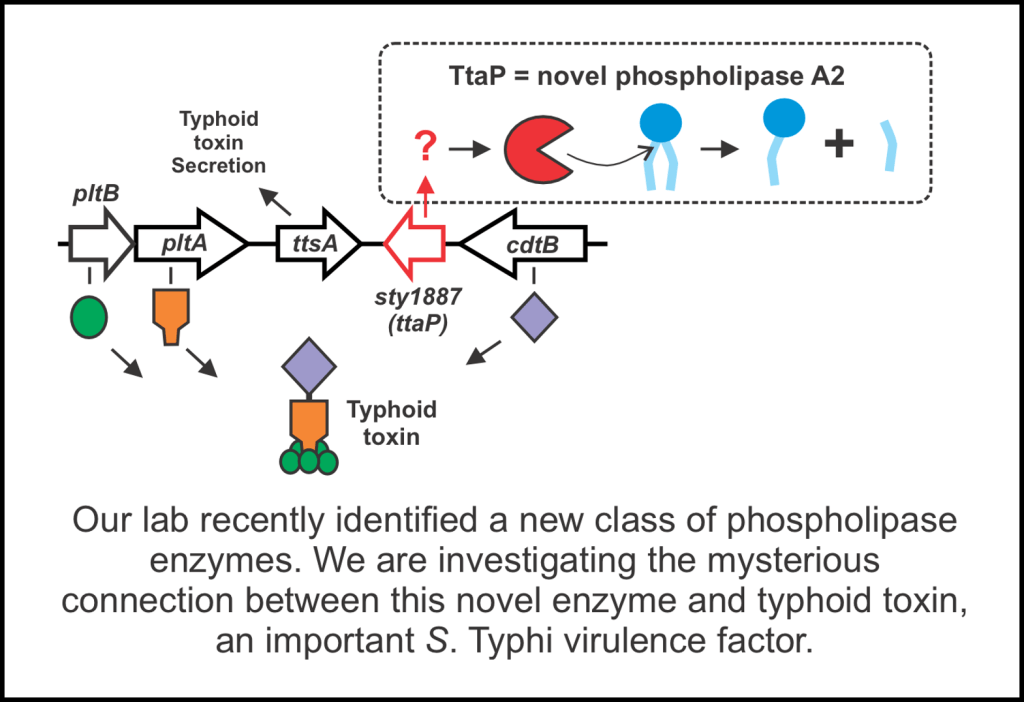
• Gartly SC, Barretto LAF, Côté AMT, Kosowan ZA, Fowler CC. A novel phospholipase A2 is a core component of the typhoid toxin genetic islet. Journal of Biological Chemistry. 2024;300(10):107758. doi:10.1016/J.JBC.2024.107758
• Chemello AJ, Fowler CC. Alternate typhoid toxin assembly evolved independently in the two Salmonella species. Miller SI, ed. mBio. 2024;15(4):e03403-23. doi:10.1128/mbio.03403-23
• Barretto LAF, Van PKT, Fowler CC. Conserved patterns of sequence diversification provide insight into the evolution of two-component systems in Enterobacteriaceae. Microbial Genomics. 2024;10(3). doi:10.1099/mgen.0.001215
• Brown PI, Ojiakor A, Chemello AJ, Fowler CC. The diverse landscape of AB5-type toxins. Engineering Microbiology. 2023;3(4):100104. doi:10.1016/j.engmic.2023.100104
• Ojiakor A, Gibbs RN, Chen Z, Gao X, Fowler CC. The evolutionary diversification of the Salmonella artAB toxin locus. Front Microbiol. 2022;13:1016438. doi:10.3389/fmicb.2022.1016438
• Barretto L, Fowler C. Identification of A Putative T6SS Immunity Islet in Salmonella Typhi. Pathogens. 2020;9(7):559. doi:10.3390/pathogens9070559
• Liu X, Chen Z, Jiao X, Jiang, X, Qiu, J, You F, Long H, Cao H, Fowler CC, Gao X. Molecular Insights into the Assembly and Functional Diversification of Typhoid Toxin. MBio. 2022;13(1):e0191621. doi:10.1128/MBIO.01916-21
• Fowler CC, Stack G, Jiao X, Lara-Tejero M, Galán JE. Alternate subunit assembly diversifies the function of a bacterial toxin. Nat Commun. 2019;10(1):3684. doi:10.1038/s41467-019-11592-0
• Fowler CC, Galán JE. Decoding a Salmonella Typhi Regulatory Network that Controls Typhoid Toxin Expression within Human Cells. Cell Host & Microbe. 2018;23(1):65-76.e6. doi:10.1016/j.chom.2017.12.001
• Fowler CC, Chang SJ, Gao X, Geiger T, Stack G, Galán JE. Emerging insights into the biology of typhoid toxin. Current Opinion in Microbiology. 2017;35:70-77. doi:10.1016/j.mib.2017.01.012
• Chen X, Taylor DW, Fowler CC, Galan JE, Wang HW, Wolin SL. An RNA Degradation Machine Sculpted by Ro Autoantigen and Noncoding RNA. Cell. 2013;153(1):166-177. doi:10.1016/j.cell.2013.02.037